This website uses cookies so that we can provide you with the best user experience possible. Cookie information is stored in your browser and performs functions such as recognising you when you return to our website and helping our team to understand which sections of the website you find most interesting and useful.
Building a Better BMI
at the Institute for Systems Biology
News

Building a Better BMI
Posted on March 20, 2023ISB researchers have constructed biological body mass index (BMI) measures that offer a more accurate representation of metabolic health and are more varied, informative and actionable than the traditional, long-used BMI equation. The work was published in the journal Nature Medicine.
The Gut Microbiome’s Supersized Role In Shaping Our Metabolome
Posted on November 10, 2022ISB researchers have shown which blood metabolites are associated with the gut microbiome, genetics, or the interplay between both. Their findings, published in the journal Nature Metabolism, have promising implications for guiding targeted therapies designed to alter the composition of the blood metabolome to improve human health.
Gut Microbiome Composition Predictive of Patient Response to Statins
Posted on May 11, 2022New ISB research shows that different patient responses to statins can be explained by the variation in the human microbiome. The findings were published in the journal Med, and suggest that microbiome monitoring could be used to help optimize personalized statin treatments.

Can You Lose Weight? Ask Your Microbiome
Posted on September 14, 2021The strongest associations with weight loss success or failure – independent of BMI – are found in the genetic capacity of the gut microbiome. These new findings open the door to diagnostic tests that can identify people likely to lose weight with healthy lifestyle changes and those who might need more drastic interventions.
Gut Microbiome Implicated in Healthy Aging and Longevity
Posted on February 18, 2021The gut microbiome is an integral component of the body, but its importance in the human aging process is unclear. ISB researchers and their collaborators have identified distinct signatures in the gut microbiome that are associated with either healthy or unhealthy aging trajectories, which in turn predict survival in a population of older individuals.
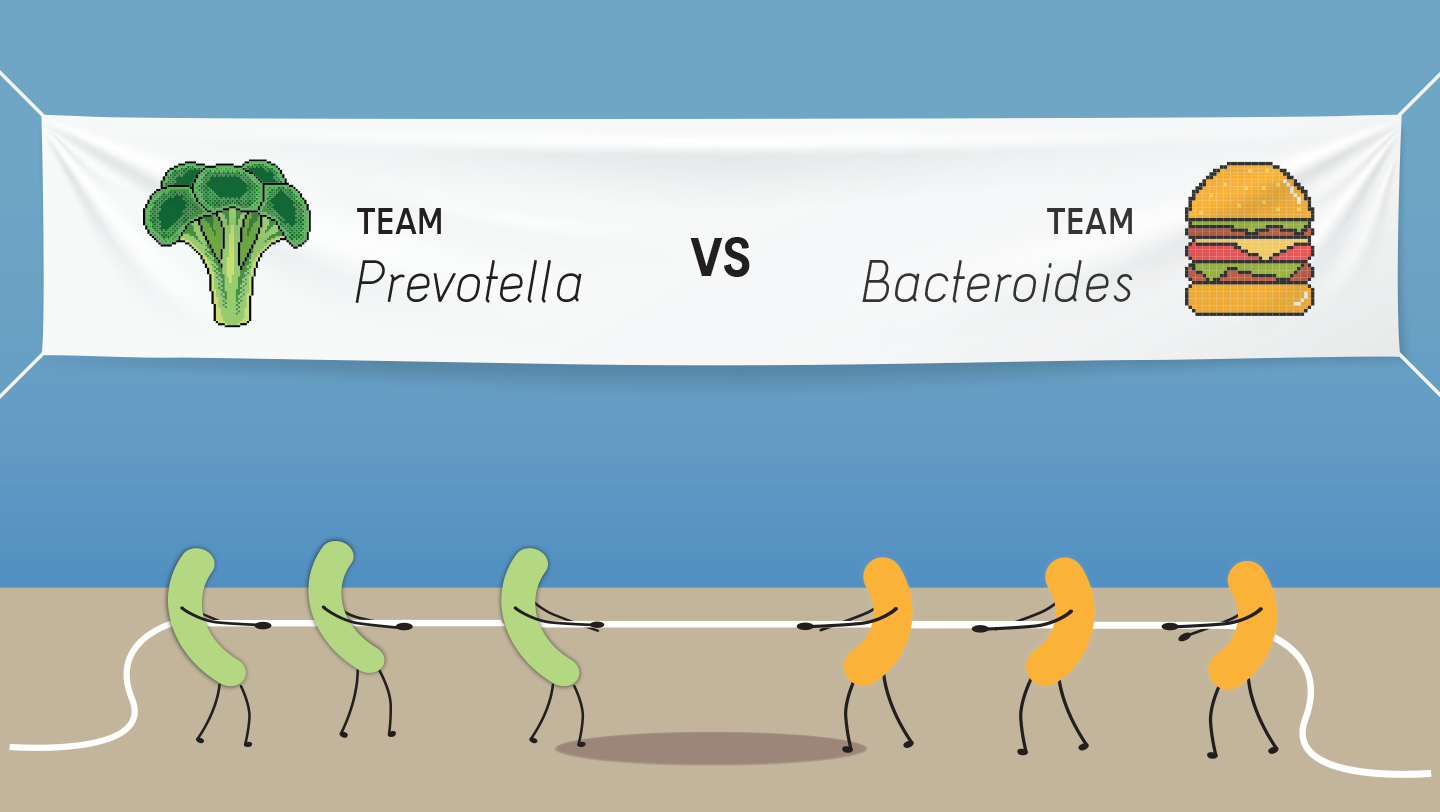
It’s ‘Either/Or’ for Two Common Gut Microbiome Genera, and Switching Teams Is Tougher Than Expected
Posted on May 29, 2020There is a dichotomy between Bacteroides- and Prevotella-dominated guts — two common gut bacterial genera — and there is a significant barrier when it comes to transitioning from one to the other.

New Modeling Tool Allows Microbiome Researchers to Map Community Ecology to Ecosystem Function
Posted on January 21, 2020A promising new open-source metabolic modeling tool provides microbiome researchers a path forward in predicting ecosystem function from community structure. News of the software package, called MICOM, was developed in part by researchers in ISB’s Gibbons Lab, and its uses were published in the journal mSystems.

Using Blood to Predict Gut Microbiome Diversity
Posted on September 2, 2019Predicting the alpha diversity of an individual’s gut microbiome is possible by examining metabolites in the blood. The robust relationship between host metabolome and gut microbiome diversity opens the door for a fast, cheap and reliable blood test to identify individuals with low gut diversity.

Seeing the microbiome through a host lens
Posted on June 7, 2019Sean recently published a commentary in the journal mSystems that outlines a vision of defining ‘microbiome health’ through a host lens: i.e. determining what exact components of the variation in the microboita influence host phenotypes. Much of the variation in the microbiome likely has nothing to do with the health state of the host, but loss/gain of critical diversity and/or functionality can have a major impact on host health. To…

Correcting Batch Effects in Microbiome Data
Posted on May 9, 2018Batch Effects in 16S Datasets Complicate Cross-Study Comparisons High-throughput data generation platforms, like mass-spectrometry, microarrays, and second-generation sequencing are susceptible to batch effects due to run-to-run variation in reagents, equipment, protocols, or personnel. Currently, batch correction methods are not commonly applied to microbiome sequencing datasets. In this paper, we compare different batch-correction methods applied to microbiome case-control studies. We introduce a model-free normalization procedure where features (i.e. bacterial taxa) in…


